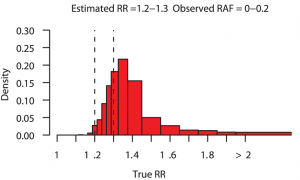
 Even though genome-wide association studies (GWAS) have identified many loci associated with complex disease, much disease heritability is still unexplained, or “missing”. But what if rather than being missing, some of the heritability was “disguised”. This is the term put forward by Chris Spencer and collegues to describe the proportion of heritability that we miss because SNPs (imperfectly) correlated to the causal variant (“tag SNPs”) are used to estimate explained heritability rather than causal variants themselves. Reassuringly, their simulations show that for the vast majority of loci detected via GWAS the risk estimated from the best tag SNP is very close to the truth. They also show that, occasionally, fine mapping of GWAS loci will identify causal variants with considerably higher risk and this is more likely if the true effect of the locus is large. The figure above, taken from their paper, shows that for estimated relative risks in the range 1.2–1.3, there is approximately a 38% chance that the true relative risk exceeds 1.4 and a 10% chance that it is over 2. The consequence of all of this for personal genomics is that disease risk could be much greater than currently thought for those individuals who, for a given disease, carry a large number of common risk variants. [CAA]
Even though genome-wide association studies (GWAS) have identified many loci associated with complex disease, much disease heritability is still unexplained, or “missing”. But what if rather than being missing, some of the heritability was “disguised”. This is the term put forward by Chris Spencer and collegues to describe the proportion of heritability that we miss because SNPs (imperfectly) correlated to the causal variant (“tag SNPs”) are used to estimate explained heritability rather than causal variants themselves. Reassuringly, their simulations show that for the vast majority of loci detected via GWAS the risk estimated from the best tag SNP is very close to the truth. They also show that, occasionally, fine mapping of GWAS loci will identify causal variants with considerably higher risk and this is more likely if the true effect of the locus is large. The figure above, taken from their paper, shows that for estimated relative risks in the range 1.2–1.3, there is approximately a 38% chance that the true relative risk exceeds 1.4 and a 10% chance that it is over 2. The consequence of all of this for personal genomics is that disease risk could be much greater than currently thought for those individuals who, for a given disease, carry a large number of common risk variants. [CAA]
↧
‘Disguised’ heritability, changes ahead to marketing of personal genomics, deciphering developmental disorders
↧